MAIN
RESEARCH
MEMBERS
PUBLICATIONS
SERVICES
COURSES
CONFERENCES
EMPLOYMENT
JOURNALS
MEETINGS
CONTACT
TOOLS
Single motif
Composite motif
Promoter fetcher
Compo
genePI
PriorsEditor
DATASETS
LOGIN
Last Update:
September 27, 2017

INTRODUCTION

PriorsEditor is a general workbench for regulatory region analysis and transcription factor binding site discovery. The program emphasizes the use of positional priors to focus the search for binding sites towards portions of DNA sequences that are considered more likely to contain true functional sites. Positional priors tracks can be constructed by manipulating, comparing and combining information about various features, including for instance phylogenetic conservation, DNase hypersensitive sites, repeat regions, nucleosome positions and other epigenetic data, physical properties of the DNA helix, distance from transcription start site or proximity to other known binding sites and many more. Data can be downloaded from the UCSC Genome Browser (or imported from file) and visualized with the graphical user interface. It is also possible to perform de novo motif discovery or motif scanning with known motif models by interfacing with locally installed programs. The results can then be visualized, analyzed and post-processed within PriorsEditor.
The software was developed by Kjetil Klepper under the supervision of professor Finn DrablÝs.
The research was supported by The National Programme for Research in Functional Genomics in Norway (FUGE) in The Research Council of Norway.
PriorsEditor is open source and free to use "as is" for academic and commercial purposes. However, no parts of the source code may be reused in its original or modified form as part of other commercial software projects without the consent of the authors.
Note that PriorsEditor can link to external programs whose use might be subject to other license restrictions.
PriorsEditor is under continuous development. For inquiries about the program, bug reports or suggestions, please contact Kjetil Klepper
DISCLAIMER
PriorsEditor is merely a tool to facilitate the construction and use of positional priors for regulatory region analysis. The authors of PriorsEditor do not have the answer to what constitutes a 'perfect prior', and it is up to the discretion of the user to manipulate and combine features as they see fit to create their own priors guided by reports in the scientific literature or their own research.
DOWNLOAD

NOTE: PriorsEditor has been superseded by a new version which has been renamed MotifLab and can be found at www.motiflab.org
PriorsEditor requires JAVA 1.6, and the latest release is version 1.0.11 (see update log)
If you have Java Web Start you can execute PriorsEditor directly by clicking on this link.
You can also download PriorsEditor for local installation.
Locally installed versions of PriorsEditor can be run from the command line (without a graphical user interface) to facilitate batch processing.
(However, it is required that you first run it once with the graphical user interface to perform necessary setup and installation steps.)
See the README file for more information.
The source code for PriorsEditor can be found here.
USER MANUAL and TUTORIALS

A comprehensive user manual will be made available in time.
Meanwhile, most questions you might have about using PriorsEditor will be answered in the following tutorials.
(Please note that the program is under continuous development and some parts of the program might look different in the latest version.)
| Tutorial #1 | General introduction. Loading and analysing data. A simple demonstration of motif scanning and post-processing |
| Tutorial #2 | Covers some additional data types and the use of operations and protocol scripts |
| Tutorial #3 | Covers the creation and use of positional priors for both pre- and post-processing |
| Tutorial #4 | A walkthrough of all the operations available in PriorsEditor |
| Tutorial #5 | Describes how to configure PriorsEditor, including adding additional feature data sources and external programs |
SCREENSHOTS

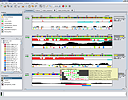
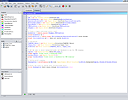
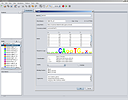
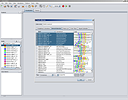
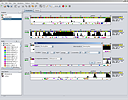
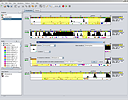
RESOURCES

PriorsEditor can interface with programs installed on the user's machine to perform motif discovery and other tasks. To use such programs, their command-line interfaces must be described in special XML-formatted configuration files which are imported into PriorsEditor from the "External Programs" dialog available under the "Tools" menu. Below is a list of premade configuration files for some popular regulatory analysis tools. Note that the configuration files listed here are also directly available within PriorsEditor from the "External Programs Repository" in the External Programs Dialog.
| BioProspector.xml | C | A Gibbs-sampling based motif discovery program | Website | |
| MotifLocator.xml | A simple motif scanning program that tend to produce lots of PWM hits | Website | ||
| MotifScanner.xml | A more advanced and discriminative motif scanning program | Website | ||
| MotifSampler.xml | A program for de novo motif discovery | Website | ||
| MEME.xml | C | P | A de novo motif discovery program that is able to use positional priors directly (from version 4.2+) | Website |
| MDscan.xml | C | Word-enumeration based motif discovery algorithm | Website | |
| Weeder.xml | C | Exhaustive word-enumeration search that allows mismatches | Website |
Programs marked with P are able to use positional priors directly. Programs marked with C require CYGWIN in order to run on Windows.
EXAMPLE PROTOCOL SCRIPTS

To use these scripts, right-click on a link and save the file on your computer. Then select "Open protocol" from the "File" menu in PriorsEditor to load the protocol. Alternatively, you can click on the link to display the protocol in the browser, select all the text and copy it to the clipboard, then choose "New protocol" in PriorsEditor and paste the text into the protocol editor. Press the "Play" button on the toolbar to execute the protocol. If you haven't specified any sequences to analyse, the Sequence Dialog will pop up and prompt you to specify your sequences (or just press "Example" to test the protocol on a set of example sequences). Note that most of these protocol scripts only work with human genomes (hg18).
| protocol1.txt | This protocol just loads lots of different features and TRANSFAC motifs and creates a few variables just for show. |
| repeatmasking.txt | Performs repeat masking of sequences and outputs the results in FASTA format. The repeats are masked with the letter 'N'. |
| conservedscan.txt | Performs PWM scanning with TRANSFAC and JASPAR matrices and filters out matches that show no phylogenetic conservation. |
| GBPscan.txt | Performs PWM scanning with TRANSFAC and JASPAR matrices and rescores the hits according to average General Binding Preference within the matching sites (see Ernst et al. below) |
CITATION

If you use PriorsEditor in your own research, please cite the following publication:
Klepper, K. and DrablÝs, F. (2010) "PriorsEditor: a tool for the creation and use of positional priors in motif discovery".
Bioinformatics, 26(17) : 2195-97 (Paper)
ACKNOWLEDGEMENTS

PriorsEditor was originally conceived as merely a tool to create positional priors tracks that could be exported to file and used by other motif discovery programs, but it has since grown to become a much more general workbench tool. In this respect, the authors of PriorsEditor would like to acknowledge the TOUCAN motif discovery workbench developed by Stein Aerts as a major source of inspiration and influence.
The background models that come bundled with PriorsEditor were obtained from the INCLUSive tools suite web site.
OTHER USEFUL REFERENCES

| Ernst J, Plasterer HL, Simon I and Bar-Joseph Z (2010) "Integrating multiple evidence sources to predict transcription factor binding in the human genome" Genome Research 20(4):526-36 PubMed |
Describes the General Binding Preference measure which is based on a linear combination of 29 features. This track is available in PriorsEditor as GBP |
| King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC (2005) "Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences" Genome Research 15(8):1051-60 PubMed Kolbe D, Taylor J, Elnitski L, Eswara P, Li J, Miller W, Hardison R, Chiaromonte F (2004) "Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat" Genome Research 14(4):700-707 PubMed |
Describes the Regulatory Potential measure which is based on a combination of phylogenetic conservation and hexamer frequency profiles (derived from known regulatory regions). This track is available in PriorsEditor as RegulatoryPotential7X |
 Bioinformatics & Gene Regulation
Bioinformatics & Gene RegulationDepartment of Cancer Research and Molecular Medicine
Norwegian University of Science and Technology
Trondheim, Norway
