SNP Highlighter
This tool highlights the locations of selected regions by drawing transparent rectangles on top of the regions that span across all the tracks in each sequence window.
The SNP Highlighter was originally intended as a visual aid to rapidly identify which TFBS that contain SNPs (and the exact affected base within the binding motif),
but it can be used to highlight other features as well besides SNPs. The image below shows an example where the locations of regions in the track "SNP141" are highlighted in pink color.
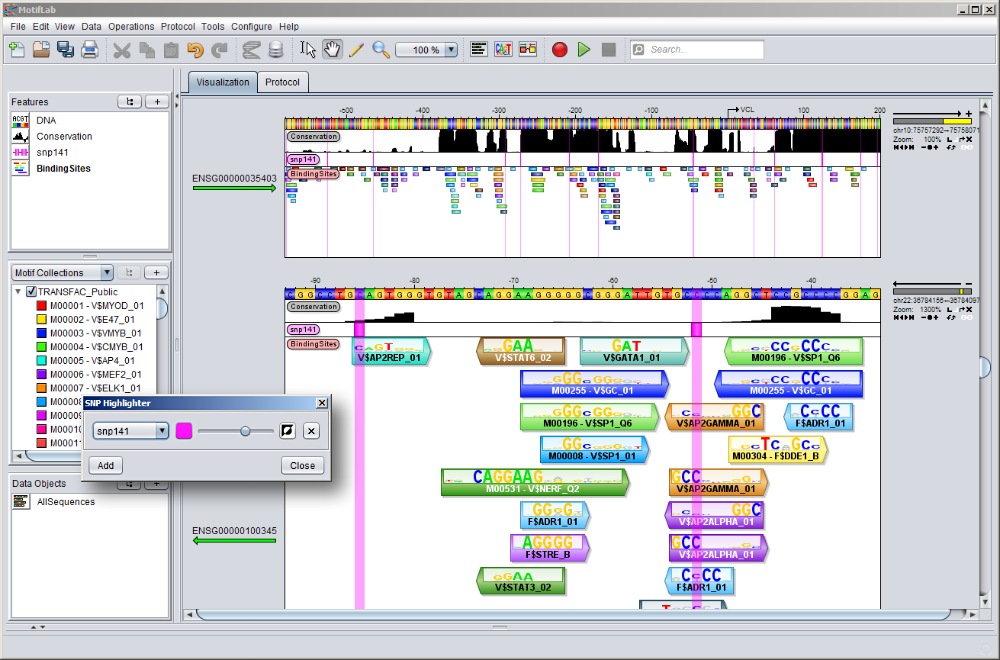
SNP Highlighter dialog
Select "SNP Highlighter" from the Tools menu to open the tool's dialog window as seen below.
The dialog will list all the active highlighters or add a new default highlighter if none are currently active.
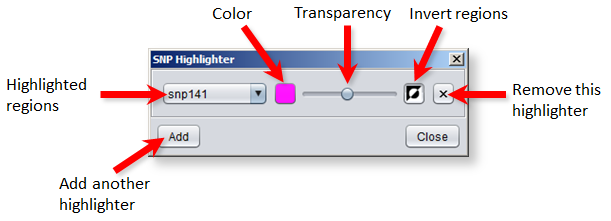
Explanations of the controls (starting from the left and going clockwise):
- The drop-down menu is used to select the track whose regions you want to highlight.
- Press the color button to select which color to use for the highlight.
- The transparency of the highlights can be adjusted with the slider.
- Pressing the "invert" button will invert the highlights so that locations outside the regions are highlighted instead.
- You can discard an active highlighter by pressing the "remove" button.
- The "Close" button will close the dialog but the current highlighters will still be active. The only way to deactivate a highlighter is to bring up the dialog again and press the remove button to discard it.
- Press the "Add" button to add more highlighters for other tracks.
Only regions that are visible within the track will be highlighted, so the tool can be used in combination with other tools or filters that dynamically change the visibility of selected regions.
However, hiding the highlighted track itself will not affect the highlights which will still be visible across the other tracks.
Each region will be highlighted individually, so if some regions are overlapping, their highlights will also be drawn on top of each other.
By adjusting the transparency to appropriate levels you can thus use highlights to visualize the density of regions across the sequence (with darker gradient colors indicating more overlapping regions).

